Table of Contents
Frap Norm
- Automatic detection of bleached and whole cell areas in pictures from FRAP experiments.
- Double or single normalization as outlined by Phair et al (2004)
Introduction
This is a basically a ROI manager to automatize measurements and calculations for FRAP analysis. It is written originally for analysis of chromatin dynamics in Arabidopsis thaliana, and uses the normalization methods outlined in : Phair et al (2004), Measurement of dynamic protein binding to chromatin in vivo using photobleaching microscopy, Methods Enzymol 375, 393-414. Both single and double normalization can be carried out.
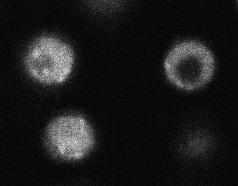
Areas are automatically outlined in three steps. First a is median filter applied. Next, the edges are detected using the Canny-deriche filter followed by hysteresis tresholding (see Edge Detection). The resulting image is displayed as guidance image for fast determination of the bleached area and the whole cell area :
The user can select the relevant ROIs with e.g. the wand tool. The plugin returns the raw measurements and if asked the normalized values for all defined areas. These can be used to determine the FRAP parameters using the appropriate software.
For more information about the use, see the help in the menu.
FRAP pictures by Stefanie Nunes Rosa
Author
Joris FA Meys (jorismeys at gmail dot com)
Installation
This plugin requires ImageEdge, which is included in the plugin Edge Detection from Thomas Boudier. Please install Edge Detection first.
Download
Download frap_norm2.jar to your plugins directory and then restart ImageJ or use the Update Menus command.
ImageJ 1.36 and Java 1.5 required