−Table of Contents
Spatial statistics 2D/3D
This plugin performs spatial statistical analysis based on distance functions F, G and H.
Author
Philippe Andrey (philippe dot andrey at versailles dot inra dot fr)
Modeling and Digital Imaging Group
Thomas Boudier  , Academia Sinica, Taipei, Taiwan.
, Academia Sinica, Taipei, Taiwan.
Features
The plugin analyses a point pattern (positions of objects of interest) distributed within a reference structure. This analysis allows in particular to assess deviation from spatial randomness, and to reveal trends for clustering (attraction) or regularity (repulsion). No edge correction is performed, as it is assumed that no point is expected outside the reference structure.
The plugin computes and reports three cumulative distribution functions (CDF):
- The G-function is the CDF of the distance between a typical point in the pattern and it nearest-neighbor.
- The F-function is the CDF of the distance between a typical position within the reference structure and its nearest point in the pattern.
- The H-function is the CDF of the distance between a typical point in the pattern and any other point.
The plugin also computes a Spatial Distribution Index (SDI), which is a normalized measure of the difference between the observed point distribution and a completely random one. The SDI is computed by comparing the empiriral CDF (F, G or H) to CDFs obtained by randomly shuffling the pattern points within the reference structure. First, the average CDF corresponding to a completely random distribution is estimated over a first set of randomly generated point patterns, conditionning on the reference structure and the number of points in the pattern. Second, the expected dispersion of CDFs around their average is estimated using a second set of randomly generated point patterns. The relative position of the empirical CDF within this range of variation is used to assign a p-value (SDI) to the pattern. If the observed points are actually randomly distributed, the SDI is uniformly distributed between 0 and 1. Further details on the method can be found in Andrey et al. 2010.
Points can be considered as hard spheres by specifying a minimum inter-point distance (hardcore distance) in the simulations of complete spatial randomness.
Usage
The plugin requires two input images, one corresponding to the binarised mask of the containing structure, one for the binarised spots contained within the structure (the point pattern to be analysed). The various parameters for the analysis are:
- The number of evaluation positions when computing F-functions
- The number of randomly generated point patterns for computing average CDF and SDI
- The minimum distance between two points (hardcore distance)
- The confidence limit of the displayed CDF confidence interval under complete spatial randomness
Illustration
P-bodies are ribonucleoprotein granules which are involved in the translational repression and degradation of eukaryotic mRNAs. Rck/p54, a RNA-binding protein required for P-body assembly in mammals, was localized in P-bodies by immuno-electron microscopy. The 10 nm gold-particles used to reveal the protein seemed to have a non-random, clustered organization, suggesting that P-bodies contain unwound RNA molecules covered with several Rck/p54 proteins. For details see Ernout-Lange et al..
Figure left: a P-body with labelled Rck/p54 proteins, magenta line indicates the manual delimitation of the P-body (reference structure), the white spots indicate the detection of the gold particles (point pattern).
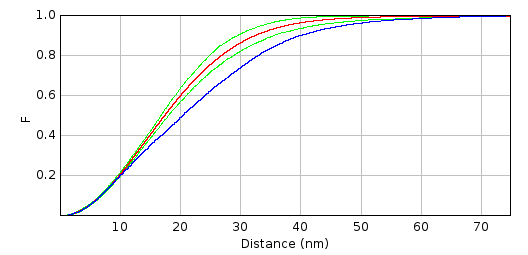
Figure right: results of analysis with F-Function. The red-line indicates the average distribution for random patterns, the green lines indicate the 95 % confidence interval and the blue line corresponds to the distribution of the observed pattern. The observed distribution lies right of the confidence interval, indicating a clustered spatial repartition.
Download
For details go to the 3D ImageJ Suite. The plugin will appear under 2D/3D Spatial Analysis in your plugins list. Note that other plugins will appear in a menu 3D in Plugins.
Citation
When using the plugin for publication, please refer to :
Andrey P, Kiêu K, Kress C, Lehmann G, Tirichine L, et al. (2010) Statistical Analysis of 3D Images Detects Regular Spatial Distributions of Centromeres and Chromocenters in Animal and Plant Nuclei. PLoS Comput Biol 6(7): e1000853. doi
J. Ollion, J. Cochennec, F. Loll, C. Escudé, T. Boudier. (2013) TANGO: A Generic Tool for High-throughput 3D Image Analysis for Studying Nuclear Organization. Bioinformatics 2013 Jul 15;29(14):1840-1. doi
License
GPL distribution (see license).
Changelog
- 25/09/2012 : First version
- 12/10/2012 : Calibration bug fixed
- 28/05/2012 : Plots start at 0. If a roi is set on a 2D image, it is the reference.