Table of Contents
AATAP
Introduction
This free code is an ImageJ plugin that allows segmentation and quantification of visceral, subcutaneaous and total abdominal adipose tissue, acquired with magnetic resonance imaging. It requires the Active contour (Snake) plugin.
Author
Daniel Kruber and Matthias Raschpichler
Description
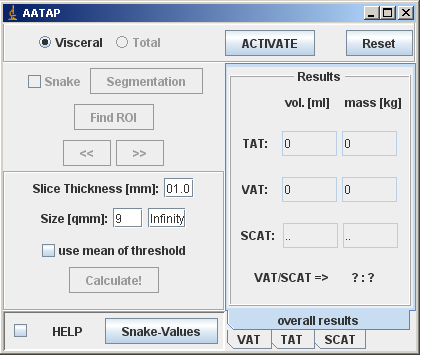
This plugin uses two steps: (1) segmentation of body from background and subcutaneous from visceral adipose tissue by two built-in Snakes; and (2) quantification of adipose tissue by counting pixels beyond an automatically adjusted threshold value. The actual volume is calculated by the equation identified fat pixel area*slice thickness*number of slices.
In detail: After loading in a stack or several images, visceral adipose tissue is quantified first. Therefore, a first RoI has to be placed manually at the external border of the ribcage on the first image, preferrably by using the freehand selection tool. This first RoI serves as an orientation for the following Snake algorithm. By checking the SEGMENTATION-button, a window allows adjusting parameters of the first Snake. Recommended Snake-parameters are provided; however, since these depend on the used MR-sequence, some individual shots might be necessary. For help, please see the Active contour (Snake) documentation. After identification of the muskulosceletal interlayer, use the FIND ROI button to anchor it on images as the new RoI.
Then, the slice thickness has to be inserted. The SIZE [qmm]-field allows definition of the minimum pixel area that will be accounted for. The next step deals with setting up an appropriate threshold value to distinguish adipose-tissue- from non-adipose-tissue-pixels. Here, the AATAP offers two choices: (1) by checking the USE MEAN THRESHOLD-function, the threshold is based on a mean histogram based on all images; otherwise (2) the threshold is based on the histogram of the currently opened, whole image. In each case, the threshold is set automatically by the a modified Isodata-algorithm that is included in the auto-threshold-function in ImageJ as default. By counting pixels beyond the threshold and inside the found RoIs, visceral adipose tissue is quantified and calculated.
By checking TOTAL, all pixel supposed to represent adipose tissue are counted. Therefore, the user repeats the above mentioned workflow identically, but locating the first RoI at the skin-background-border on the first image. Finally, after calculating total abdominal adipose tissue, subcutaneous adipose tissue is calculated by the equation total abdominal adipose tissue - visceral adipose tissue.
The right column provides overall results as well as the results for each slice.
Installation
Download aatap_.zip and extract to a new folder (for instance: AATAP) in the ImageJ plugins folder. Then, compile the AATAP_ - file using Plugins/Compile and Run. There will be a new “AATAP” folder with a new “AATAP” command in the Plugins menu the next time you restart ImageJ.
License
The program is open source; you can redistribute is and/or modify it under the terms of the GNU General public License.
Changelog
We truly hope that this program is useful, but WITHOUT ANY WARRANTY. Feel free to improve or adjust it for your needs.