plugin:segmentation:jslic:start
Table of Contents
jSLIC - superpixels
We made a Java-based open source implementation jSLIC - the superpixel clustering with better performance than the original Simple Linear Iterative Clustering. For more info, please see Fiji wiki.
How to use the plugin
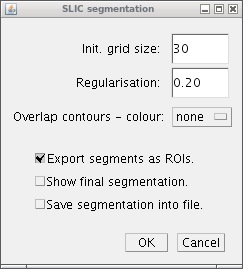 As you can see the Interface to the plugin contains a parameters for superpixel configuration and also its final visualisation.
As you can see the Interface to the plugin contains a parameters for superpixel configuration and also its final visualisation.
Configuration
For the configuration there are only two parameters to be set:
- Init. grid size - in general it can be seen as an average superpixels size.
- Regularisation - influence the compactness of estimated superpixels. The range is from 0 (very elastic superpixels) to 1 (superpixels are nearly squares). Experimentally, we set as optimal value 0.2 for most cases.
Visualisation
To show of handle segmentation results we presented a few approaches:
- Overlap contours - simply draw the contours on resulting superpixels into the image by chosen colour.
- Export segments as ROIs - all superpixels are exported as polygons into the ROI Manager.
- Show final segmentation - add one more stack and fill each superpixel by a random colour.
- Save segmentation into file - export the superpixel segmentation into a text file as segmentation matrix with labels.
References
Borovec, J., & Kybic, J. (2014). jSLIC : superpixels in ImageJ. Computer Vision Winter Workshop. Praha.
plugin/segmentation/jslic/start.txt · Last modified: 2019/04/12 13:13 by 127.0.0.1


