Table of Contents
3D Segmentation
This plugin implements various algorithms to segment 3D images.
Author
Thomas Boudier
Features
Several algorithms for segmentation are proposed :
- 3D hysteresis thresholding with two thresholds (see 2D hysteresis for explanation).
- 3D simple segmentation with thresholding to label 3D objects (similar to 3D objects counter).
- 3D iterative thresholding (find optimal threshold for each object).
- 3D spot segmentation with various local threshold estimations.
- 3D Maxima Finder (with noise parameter)
- 3D seeds-based watershed with automatic local maxima detection for seeds.
3D Iterative Thresholding
The plugin will basically test all thresholds and detect objects for all thresholds, it will then try to build a lineage of the objects detected, linking them from one threshold to the next threshold, taking possible splits into account. So different objects can be segmented with different thresholds, the plugin allows various criteria to pick the best threshold :
- Elongation : the threshold leading to the most round object is chosen (minimal elongation).
- Volume : the threshold leading to the largest object.
- MSER : the threshold where the volume of the object is most stable (minimal variation).
- Edges : the threshold where the objects has maximal edges (see edge plugin)
The other parameters are related to the minimal and maximal volumes of the objects. The thresholds tested can be tuned with 3 options with the value parameter :
- Step : threshold are tested each step value.
- Kmeans : histogram is analysed and clustered into value classes using a KMeans algorithm.
- Volume : a constant number of pixels between two thresholds using value thresholds.
For 8-bits images it is recommended to use the method Step with value between 1 and 5. For 16-bits images try Step with values between 5 and 100 depending on the dynamic of your data. Note than the more threshold tested the more memory used. In order not to test low thresholds you can specify to start with the mean value of the image as the lowest threshold or specify manually the lowest threshold to start with. The image can be filtered before thresholding with a 3D median filter with radii proportional to the minimal volume. The contrast refers to the range of thresholds where the object exists, noise or very faint objects may have very low contrast as opposed to very contrasted object.
 Fig. 1: Iterative thresholding using different criteria, bottom left elongation, top right volume and bottom right MSER.
Fig. 1: Iterative thresholding using different criteria, bottom left elongation, top right volume and bottom right MSER.
Testing all thresholds may lead to objects being divided into smaller objects for high thresholds. For instance touching cells may result in close nuclei, at low contrast and low threshold the two nuclei may seem like touching and form only one object, however at high threshold and contrast two separate objects are being seen.
 Fig. 1: Dividing objects with thresholds, top left raw image with high brightness, top right raw image with adjusted contrast to distinguish the dividing nuclei, bottom left first channel of Iterative thresholding showing brighter and smaller objects, bottom right second channel of Iterative thresholding showing merged nuclei for lower threshold.
Fig. 1: Dividing objects with thresholds, top left raw image with high brightness, top right raw image with adjusted contrast to distinguish the dividing nuclei, bottom left first channel of Iterative thresholding showing brighter and smaller objects, bottom right second channel of Iterative thresholding showing merged nuclei for lower threshold.
If you find this plugin useful for your work, please cite this paper and refer to the 3D Image Suite page : A generic classification-based method for segmentation of nuclei in 3D images of early embryos
3D Spot Segmentation
The plugin works with two images, one containing the seeds of the objects, that can be obtained from local maxima (see 3D filters), the other image containing signal data. The program computes a local threshold around each seeds and cluster voxels with values higher than the local threshold computed. A plugin 3D maxima Finder is also available to compute the seeds.
Three methods are available for computing the value of the local threshold and 3 methods for clustering are also proposed. The option watershed can be chosen to avoid merging of close spots.
A tutorial is also avalaible : 3D Spot Segmentation Manual.
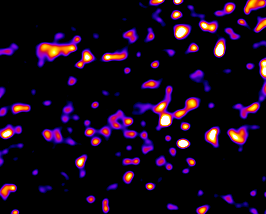

 Fig. 3: Spot segmentation. Left slide of a 3D raw image with crowded objects with different intensities. Middle the zones around each detected local maxima, comuted using watershed. Right the final segmentation of the objects.
Fig. 3: Spot segmentation. Left slide of a 3D raw image with crowded objects with different intensities. Middle the zones around each detected local maxima, comuted using watershed. Right the final segmentation of the objects.
3D Watershed
The plugin works with two images, one containing the seeds of the objects, that can be obtained from local maxima (see 3D filters), the other image containing signal data. A first threshold1 is used for seeds (only seeds with value > threshold1 will be used). A second threshold is used to cluster voxels with values > threshold2. In this implementation voxels are clustered to seeds in descending order of voxel values. An alternative implementation is available in Fiji classic watershed.
Two plugins 3D splitting and 3D Voronoi are also available, more details in this brief tutorial.
Download
For details go to the 3D ImageJ Suite.
Citation
When using the 3D Segmentation plugins for publication, please cite :
J. Ollion, J. Cochennec, F. Loll, C. Escudé, T. Boudier. (2013) TANGO: A Generic Tool for High-throughput 3D Image Analysis for Studying Nuclear Organization. Bioinformatics 2013 Jul 15;29(14):1840-1. doi
License
GPL distribution (see license). Sources for plugins are available freely. Sources for core are available on request.
Changelog
- 24/10/2013 V2.7 : new Iterative Thresholding; 32-bits segmentation and labelling for large number of objects (>65 535)
- 21/03/2014 V2.71: bug fix in Segment3D
- 25/05/2015 V3.2 : improved watershed (especially for flat regions)
- 20/08/2015 V3.4 : redesigned Watershed, record Iterative Thresholding
- 22/08/2016 V3.83: watershed improved