−Table of Contents
Confined Displacement Algorithm Determines True and Random Colocalization
Introduction
The quantification of colocalizing signals in multi-channel fluorescence microscopy images depends on the reliable segmentation of corresponding regions of interest (ROIs), on the selection of appropriate colocalization coefficients, and on a robust statistical criterion to discriminate true from random colocalization. Our plugin contains the confined displacement algorithm (CDA) based on image correlation spectroscopy in combination with Manders Colocalization Coefficients M1ROI and M2ROI to quantify true and random colocalization of a given florescence pattern. Existing algorithms based on block scrambling exaggerate the randomization of fluorescent pattern with resulting inappropriately narrow probability density functions (PDFs) and false significance of true colocalization in terms of p values. Our CDA algorithm approach detects true colocalization of specific fluorescence patterns down to sub-cellular levels.
For a more detailed description of the algorithm, please refer to Ramirez O, Garcia A, Rojas R, Couve A, and Härtel S (2010) Confined Displacement Algorithm Determines True and Random Colocalization in Fluorescence Microscopy. Journal of Microscopy. ( our web site).
Authors
Ramirez O, Garcia A, Rojas R, Couve A, Härtel S, Díaz-Espinoza D, Osorio-Reich M
Description
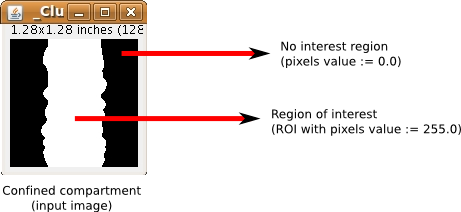
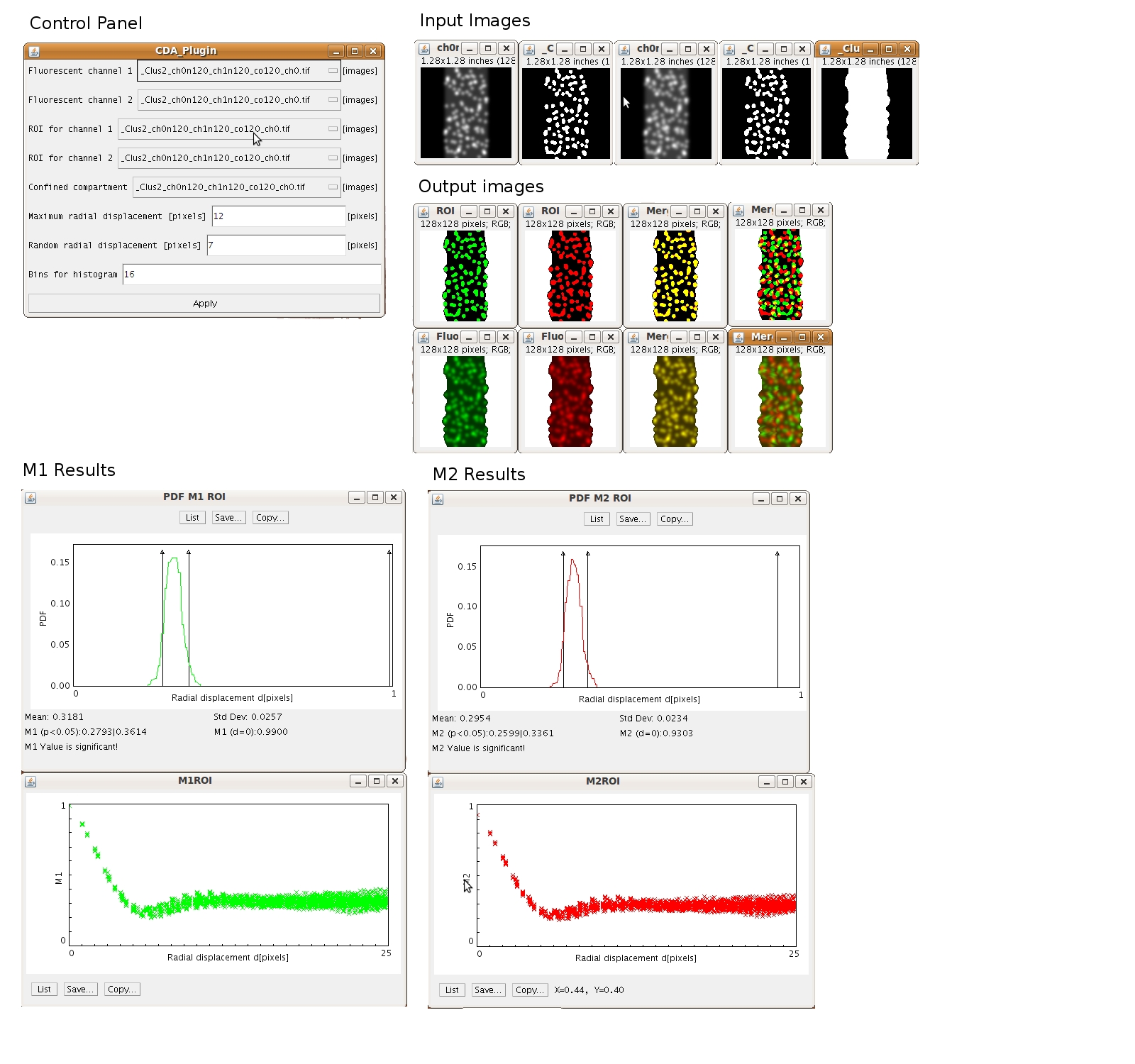
This plugin calculates modified Manders coefficients, M1ROI and M2ROI, in combination with image correlation techniques in order to assess true and random colocalization statistically. To generate random scenarios for the given ROIs, one channel and its corresponding image mask is radially shifted (on interval [0, d_final]) relative to the second channel. This two-dimensional image correlation technique calculates M1/2ROI(d), as a function of pixel displacements. As the displacement increases, M1/2ROI(d) approach values that represent random scenarios for a given signal distribution. The interval of M1/2ROI between [d_random, d_final] will be taken as the random scenarios on which the statistical significance test will be applied. In order to maintain reliable characterization of random colocalization, the radial displacements are confined to the previously defined cellular compartments. Pixels which are shifted out of these compartments are subsequently inserted on the opposite side. Thereby, CDA plots can reliably represent random scenarios by maintaining signal density, mean signal density and signal pattern. PDFs are calculated using these random values of M1/2ROI(d) to test if the colocalization coefficients that were calculated at the original image position [ M1/2ROI (d=0) ] are statistically different from a random population in terms of p values.
Note, M1/2ROI ∈ [0.0, 1.0], with 1.0 representing 100% colocalization and 0.0 representing 0% colocalization.
Installation
Download .jar file and save in the Plugins directory of ImageJ. Restart ImageJ.
Download
Getting started
Before running the plugin, open the 5 images in ImageJ being used for analysis. Then, click on the ImageJ Plugins Menu → CDA → Confined Displacement Algorithm. Choose the appropriate images in the drop down menu and enter parameters, as follows:
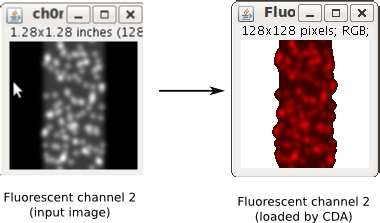
- one image for the green channel (fluorescent channel 1). This channel will be radially shifted together with its ROI (see Description).
- one image for the red channel (fluorescent channel 2).
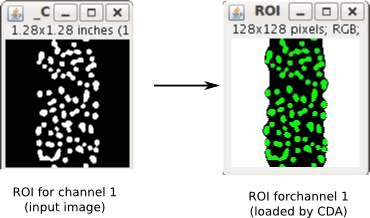
- one image for the segmented binary region of interest (ROI for channel 1). This ROI will be radially shifted with its fluorescent channel (see Description).
- one image for the segmented binary region of interest (ROI for channel 2).
- one image with a ROI on which to confine the colocalization analysis (Confined compartment).
- enter the maximum pixels to shift channel 1 (maximum radial displacement).
- enter the radial displacement for the statistical significance test (random radial displacement).
- finally enter the number of bins for the gaussian distribution (bins for histogram).
Note 1: If you upload a multichannel image for the fluorescent channel, the plugin will only use the first channel. ROI images must only have pixels values 0 and 255.
Note 2: maximum radial displacement > random radial displacement.
Changelog
2010/02/03: Beta version.
2011/05/23: Eliminate use of obsolete PlotWindow class for displaying histogram & text results.
About us
The Laboratory of Scientific Image Analysis (SCIAN-Lab) develops mathematical tools and computational algorithms to access dynamic, morphologic, and topologic features in experimental systems with a biophysical, biological, or medical background. High speed confocal microscopy in combination with image processing routines unravels the interplay between structure and function in lipid model systems and phenomena on sub-cellular, cellular, and supra-cellular levels in the field of developmental biology, neurobiology, or membrane biophysics. For further information, please visit our website.