−Table of Contents
TAPAS Tutorial : Measurement
In this tutorial we will learn some basic analysis functions of TAPAS. We will learn how to use the measurement module. Please check you understand the basics of TAPAS and have a look to this tutorial on Input/Output. You can also check this tutorial on segmentation with TAPAS.
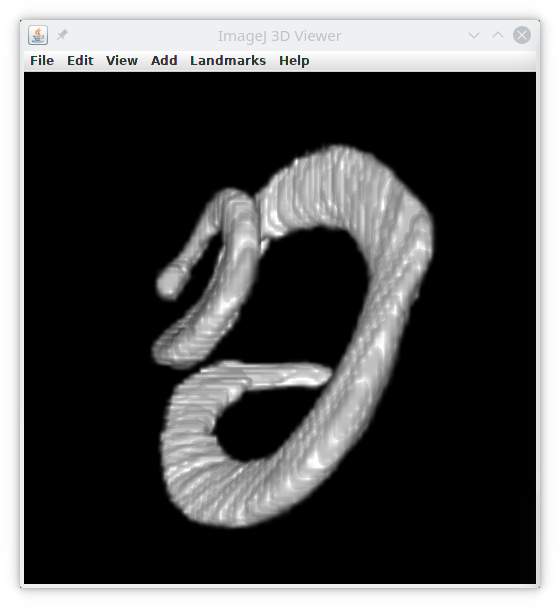
In this tutorial we will use the BatCochleaVolume and T1-head images from ImageJ, download it from Open Samples.
Basic usage
TAPAS is focusing on 3D measurements, while most of 3D measurements are still valid in 2D, some 3D measurements should be used with caution with 2D data.
The basic module for measurement is measurement, it requires a labeled image, with all individual objects having their own value. If your image is not labeled yet, please refer to this tutorial on segmentation.
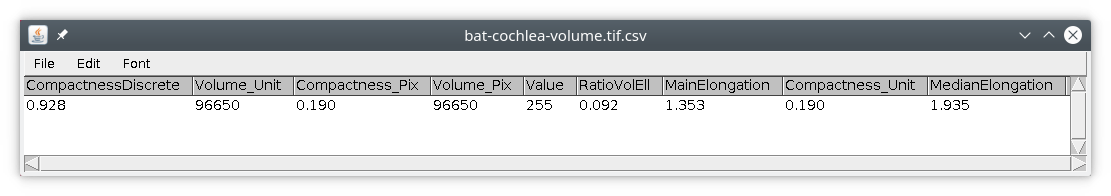
The module will measure the individual object and save the results in a csv file, that can be read with a spread sheet editor or directly into ImageJ. The available measurements available are :
- volume, in pixels and units
- area, in pixels and units
- centroid, the coordinates of the center of the object, in pixels and units
- compactness, the ratio between volume and area, no unit
- ellipsoid, the fitting with an ellipsoid, with main elongation (see this plugin for details)
- DC, distances from the center to the contour
Here is how to use this module with TAPAS :
// load the data process:input // do the measurement of objects in the image // save results in temporary file, here user home directory // as output file for results // we use the name of the image being processed // followed by -results.csv process:measurement dir:?home? file:?name?-results.csv list:volume,compactness,ellipsoid // attach results to original file process:attach dir:?home? file:?name?-results.csv
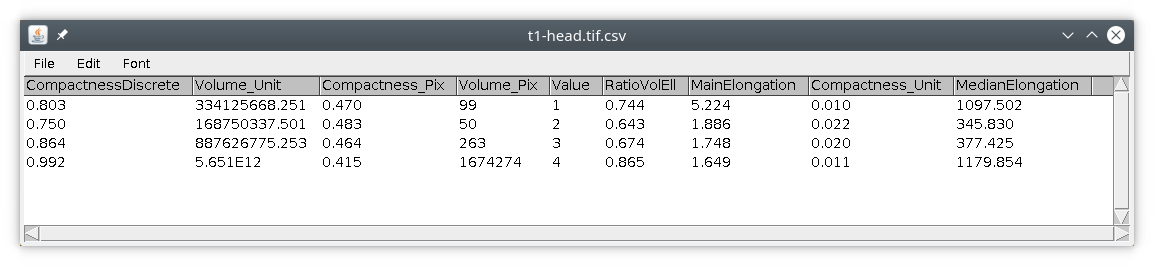
If we run this module with the bat-cochlea image, we simply obtain this results table :
Advanced usage
In this section we will write a full processing pipeline and see how to focus on only the biggest object in the image and get rid of all smaller objects.
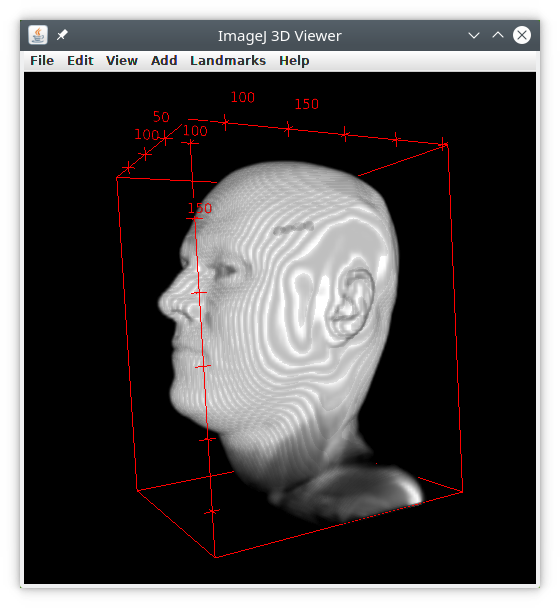
We will use t1-head as example.
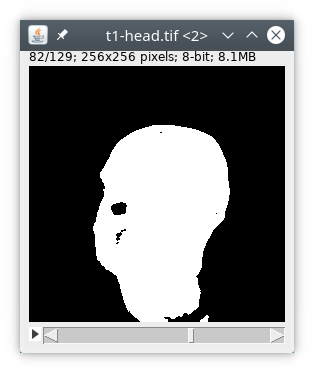
We will first filter the image by a 3D median filter to homogenize signal and then use an auto thresholding to detect the head. The corresponding text is then :
// load the data process:input // median filter process:filters filter:median radxy:2 radz:2 // automatic thresholding process:autoThreshold method:Triangle
We will then label the binary image to detect the objects inside the image, and then perform the measurements of these objects. The corresponding text is then :
// label the objects // min volume in pixels // use unit:yes when minVolume is in unit process:label minVolume:50 // do the measurement of objects in the image // save results in temporary file, here user home directory process:measurement dir:?home? file:?name?-results.csv list:volume,compactness,ellipsoid
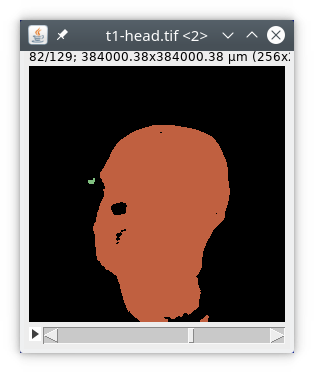
If we look at the results table we can see there are actually four objects inside the image.
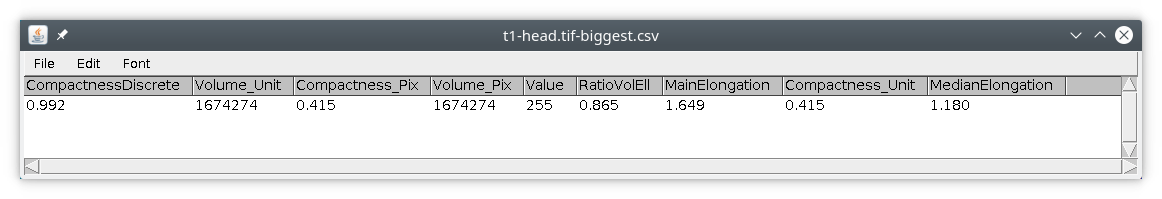
So we could increase the parameter minVolume in the module label. We can also use another module simply called biggest. The final processing pipeline will then look like this :
// load the data process:input // median filter process:filters filter:median radxy:2 radz:2 // automatic thresholding process:autoThreshold method:Triangle // label the objects // min volume in pixels // use unit:yes when minVolume is in unit process:label minVolume:50 // keep only the biggest object process:biggest // do the measurement of objects in the image // save results in temporary file, here user home directory process:measurement dir:?home? file:?name?-biggest.csv list:volume,compactness,ellipsoid // attach results to original file process:attach dir:?home? file:?name?-biggest.csv
We then obtain an image with only the biggest object, the head, and only one object in measurements.