Table of Contents
TAPAS Tutorial : 2D Measurements with AnalyzeParticles
In this tutorial we will learn how to use analysis features of ImageJ within TAPAS. We will learn how to use the analyseParticles module. Please make sure you understand the basics of TAPAS and have a look to this tutorial on Input/Output. You can also check this tutorial on segmentation with TAPAS, tutorial on measurement with TAPAS and this tutorial on quantification.
In this tutorial we will use the Blobs image from ImageJ, download it from Open Samples. We just removed the inverted look-up table by selecting LookUp Table Grays.
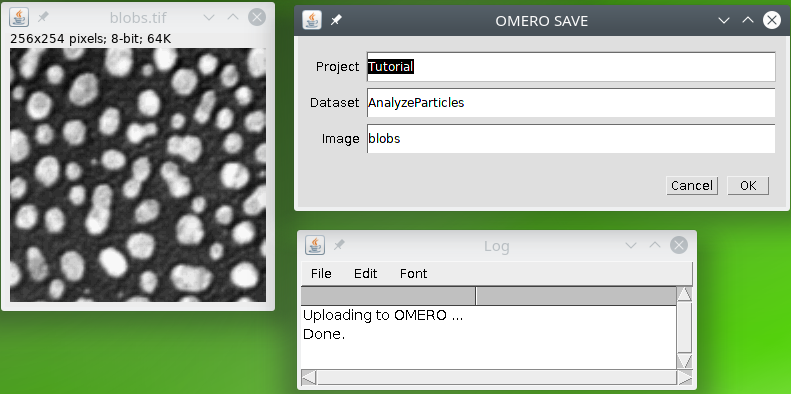
We can upload the data to Omero if you have it or simply copy the files into our directory structure (see here for details).
Basic usage
In this tutorial we will learn how to use the AnalyzeParticles module from TAPAS. This module wraps up the functionalities of the Analyze Particles function in ImageJ. This module will take a binary image as input and will output a labeled (a.k.a. segmented), image, where objects are individually identified. A results table file will also be exported.
The first thing is to specify which objects to detect, we need to specify a minimum and maximum size, as well as a minimum and maximum circularity. The defaults values are 0 for minimum size, infinity for maximum size (actually specified as negative value -1), minimum and maximum values for circularity are 0 and 1. The default values will allow all objects to be detected. You can restrict what kind of objects you want to be detected, e.g. only big objects by setting a minimum size, or only round objects by specifying a minimum circularity.
You need also to specify the measurements you want to perform on the objects as a list of measurements. Available measurements are : area, perimeter, centroid, ellipse (fitting), shape (descriptors) and Feret (diameter).
The code for basic usage is then :
// Analyze particles, results are saved // in a file called results.csv by default process:analyzeParticles dir:?ij? file:myResults.csv list:centroid,ellipse,feret,shape
On our example before analysis, we need to filter the image a bit to homogenize intensities, then apply a threshold. The full code is then :
// input data process:input // filter with median filter radius 2in xy process:filters radxy:2 filter:median // we threshold the image with Otsu thresholding process:autoThreshold method:OTSU // Analyze particles, results are saved // in a file called results.csv by default process:analyzeParticles dir:?ij? file:myResults.csv list:centroid,ellipse,feret,shape // to check the results of previous module // we simply display the image process:show
We then obtain the following labeled image :
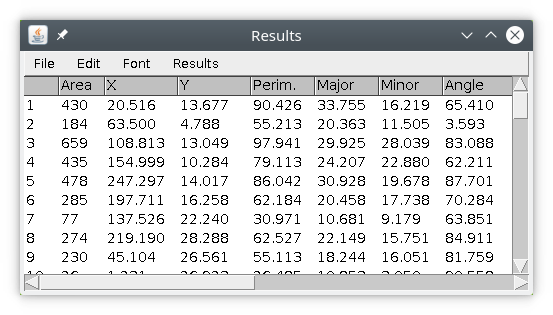
and the following results table :
Advanced usage
We can also use the parameter excludeEdges:yes to remove particles touching the edges.
The code to remove particles on edges is then :
// Analyze particles, results are saved // in a file called results.csv by default process:analyzeParticles dir:?ij? file:myResults.csv excludeEdges:yes list:centroid,ellipse,feret,shape
We obtained this segmented images where particles touching the edges of the image are removed :
Here a simple example where we keep only bigger round particles, by specifying a minimum circularity of 0.9 and a minimum size of 10. By default, sizes are in pixels, if you want to specify sizes in unit just add the parameter unit:yes.
process:analyzeParticles dir:?home? excludeEdges:yes minSize:10 minCirc:0.9 list:centroid,ellipse,feret,shape
Saving the results
We will finally save the results of segmentation and measurements in the Omero database or in the same folder as the input if you do not have Omero. We will then use two output commands to save the segmented image, and attach to link the measurements results file to the original image. We will also delete the temporary results file using the command delete.
// output the segmented image process:output name:?name?-seg // attach the results file // to the original image process:attach dir:?home? file:results.csv // delete temporary results file process:delete dir:?home? file:results.csv
The full code is then :
// input, filter and thresholding process:input process:filters radxy:2 filter:median process:autoThreshold method:OTSU // segmentation and measurements process:analyzeParticles dir:?home? file:results.csv list:centroid,ellipse,feret,shape // output and delete temporary file process:output name:?name?-seg process:attach dir:?home? file:results.csv process:delete dir:?home? file:results.csv